 |
| Research
| CYTOKININ
SIGNALING PATHWAY
  Supported
by the NSF IBN Program and the NIH Supported
by the NSF IBN Program and the NIH
Summary |
|
|
Cytokinins
are essential plant hormones that control cell division,
shoot meristem initia-tion, leaf and root differentiation,
chloroplast biogenesis, stress tolerance, and senescence.
Together with auxin, another plant hormone, cytokinins
can reprogram terminally differen-tiated leaf cells
into stem cells and support shoot regeneration indefinitely
in plant tissue culture (1, 2). Thus, cytokinins are
master regulators of plant growth and development, which
are highly plastic and adaptive, as well as remarkably
resilient and perpetual. Research interest in the signaling
pathways activated by cytokinins has increased recently
because of new information arising from studies of Arabidopsis
and the completion of its genome sequence. However,
the importance of this pathway is given additional weight
because it represents two-component signaling, a canonical
mechanism that mediates diverse biological responses
in many taxa. The specific Cytokinin Signaling Pathway
(3) details the pathway as it has been elucidated in
Arabidopsis; the canonical Cytokinin Signaling Pathway
presents the general view (4). |
| |
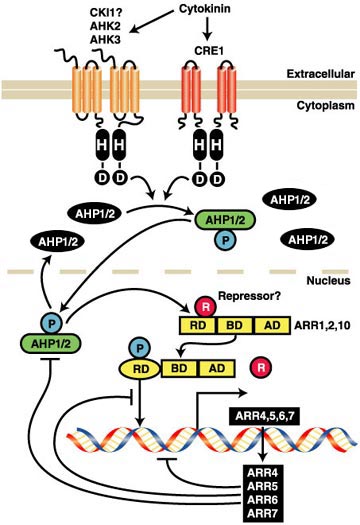 |
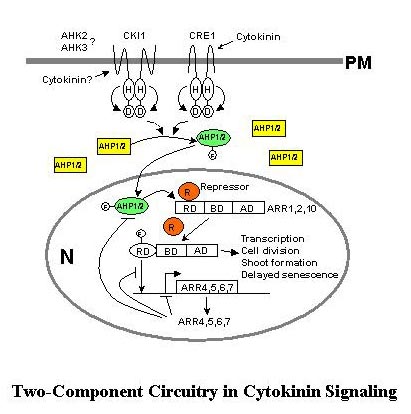
In the Arabidopsis
cytokinin signal transduction pathway, hybrid histidine
protein kinases (AHKs) serve as cytokinin receptors
and histidine phosphotransfer proteins (AHPs) transmit
the signal from AHKs to nuclear response regulators
(ARRs), which can activate or repress transcription
(5–10). Similar components are also found in maize,
suggesting a conservation of the cytokinin signaling
mechanism in plants (11). There are four major steps
to cytokinin signaling: AHK sensing and signaling, AHP
nuclear translocation, ARR transcription activation,
and a negative feedback loop through cytokinin-inducible
ARR gene products (Fig. 1). Analyses of mutants and
transgenic tissues and plants support the importance
of this central signaling pathway in diverse cytokinin
responses (5–10). The multistep two-component phosphorelay
mechanism found in Arabidopsis is reminiscent of the
bacterial two-component signaling system (12), but it
is linked by AHPs, which shuttle from the cytoplasm
to the nucleus in a cytokinin-dependent manner (6).
Although conserved motifs for two-component phosphorelay
systems have been identified in plant hormone ethylene
receptors (13), phytochrome photoreceptors (14), and
a putative osmosensor (15), until recently the importance
of histidine protein kinase activity and phosphorelay
had not been demonstrated in plant cells. Functional
analyses of AHKs, AHPs, and ARRs in Escherichia coli,
yeasts, plants, and a leaf protoplast system, and protein-protein
interactions in yeast two-hybrid assays, have provided
compelling evidence for the importance of multistep
two-component phosphorelay in cytokinin signaling (5–10,
16–18). |
| |
|
| |
In Arabidopsis,
at least three genes encode cytokinin receptors: AHK4
[also known as CYTOKININ RESPONSE 1 (CRE1) and WOODEN
LEG (WOL)], AHK2, and AHK3 (7, 19, 20). Other Arabidopsis
histidine protein kinases, cytokinin independent 1 (CKI1)
and CKI2 (also known as AHK5), can also activate cytokinin
responses in the absence of exogenously added cytokinin
(5, 6). Quantitative transcription analyses based on
cytokinin-inducible ARR6-LUC reporter gene activity
suggest that CKI1 and AHKs act through different cytokinin
perception mechanisms. CKI1 is constitutively active,
but AHK4, AHK2, and AHK3 require extracellular cytokinin
for their activation (6). The function of AHK4 has been
thoroughly demonstrated by direct cytokinin binding
(21) and by the isolation of cre1 and wol mutants that
exhibit defects in cytokinin-mediated shoot induction
from callus and root vascular morphogenesis, respectively
(7, 19). The lack of shoot phenotypes in cre1 and wol
suggests that the functions of AHK2 and AHK3 may overlap
with that of AHK4 (20). Further analyses of cellular
expression patterns, cytokinin binding, and chimeric
AHKs with swapped domains should clarify the underlying
mechanism of each AHK action in cytokinin signaling.
|
| |
|
| |
The analysis
of fusions between green fluorescent protein (GFP) and
AHP (AHP-GFP) has provided the first visual, in vivo
evidence that AHP1 and AHP2 are translocated into the
nucleus in a cytokinin-dependent manner (6). In Arabidopsis,
there are more AHKs, ARRs, and related proteins than
there are AHPs (18, 22),suggesting that multiple two-component
signaling pathways may share AHPs (6, 10). The cytokinin
pathway does not follow the established eukaryotic histidine
protein kinase and mitogen-activated protein kinase
(MAPK) cascade paradigm (23), but rather integrates
multiple AHK activities to common AHPs, which then modulate
distinct ARRs in the nucleus (6). |
| |
|
| |
The B-type ARR
transcription activators (ARR1, ARR2, and ARR10) carry
MYB-like domains for DNA binding and a glutamine (Q)-rich
domain for transcriptional activation (24, 25), and
they activate cytokinin-responsive ARR6 transcription
(6, 8). These activators appear to be the evolutionary
products of domain shuffling, with ancestral modules
originating from both prokaryotic and eukaryotic heritage.
Mutation in the conserved aspartate residue of ARR2
does not abolish its function as a transcription activator
for a cytokinin early-response gene ARR6 promoter, suggesting
that phospho-rylation may not intrinsically activate
the transcription factor (Fig. 1) (6). Consistently,
deletion of the receiver domain of ARR1 results in higher
transcription activity in plant cells and constitutive
cytokinin phenotypes in transgenic plants (8, 24). Thus,
phosphorylation of ARR1 and ARR2 likely eliminates negative
regulation (Fig. 1). Ectopic expression in transgenic
Arabidopsis of ARR2, one of the rate-limiting transcription
factors in the response to cytokinin, is sufficient
to mimic cytokinin in promoting shoot meristem proliferation
and leaf differentiation, and in delaying leaf senescence
(6). The lack of striking phenotypes in the arr1 mutant
indicates that multiple B-type ARRs may serve similar
functions (6, 8). Determining the target genes of these
transcription factors using microarrays will add new
insight into the molecular basis of cytokinin actions.
|
| |
|
| |
The products of
the cytokinin-inducible A-type ARR4, ARR5, ARR6, and
ARR7 genes inhibit transcription, which could mediate
a negative feedback loop that controls the tran-sient
induction of cytokinin primary response genes and allows
resetting and/or fine-tuning of the physiological state
of the cells (Fig. 1) (6, 16). Although the B-type ARRs
with transcriptional activation activities are likely
the major regulators of a broad spectrum of cytokinin
target genes (26), the A-type ARRs could also contribute
to the outputs of cytokinin signaling through protein-protein
interactions (16, 17). Two-component elements could
potentially be regulated by signals other than cytokinin
and provide a cross-talk mechanism in plant signaling
networks. For instance, expression of some ARRs is regulated
by stress (27) and sugar signals (28). ARR4 also interacts
with phytochrome B and modulates light signaling (29).
Thus, two-component elements could serve as the molecular
links in a complex plant signal transduction network
that sensitively integrates central growth signals such
as plant hormones, sugars, light, and other environmental
cues. |
| |
|
| |
The expression
analysis of CYCLIN D (30) and an ARR5::GUS transgene
(31)inArabidopsis has shown that root and shoot meristems
are major sites of cytokinin actions. However, cytokinin
responses can also occur in other cell types (6, 31).
This broad cellular competence to cytokinin responses
may explain the plasticity of plant development. The
emerging short cytokinin signaling circuit could represent
a conserved core signaling pathway in different cell
types in response to cytokinin. However, additional
cell type–specific components are likely to play important
roles for cytokinin responses in different cell types
and tissues, for example, in dividing and nondividing
cells. Elucidation of the expression patterns and subcellular
localization of AHKs, AHPs, and ARRs will contribute
to a better understanding of their unique or overlapping
roles in cytokinin responses and in other two-component
signaling pathways in plants. The major challenge is
to determine how a conserved cytokinin signal transduction
pathway influences cell cycle, leaf senescence, shoot
initiation, and leaf patterning in different cell types
at various developmental stages. |
| |
|
| |
The completion
of the Arabidopsis genome sequence has revealed 54 genes
encoding puta-tive AHKs, AHPs, ARRs, and related proteins,
suggesting a substantial involvement of this signaling
mechanism in many facets of plant cell regulation (17,
18, 32). The development of the Arabidopsis protoplast
system has enabled a high-throughput functional genomic
analysis of the two-component regulators (6). Because
pronounced redundancy in the Arabidopsis genome is evident
(18, 32), cellular analyses of the two-component elements
would complement the characterization of a large number
of insertion mutants that may not display overt phenotypes.
Genetic, genomic, and biochemical experiments will elucidate
the details in cytokinin perception, protein-protein
interactions, and target gene expression essential in
cytokinin signaling. |
|
|
| Signaling
Pathway Illustrations
| Legend: Colors indicate the
localization of a component. (This is, of course, a
rough guide for orientation on the map, as many components
change locations during signaling.) 
To see more detailed information about each
component, click
here. |
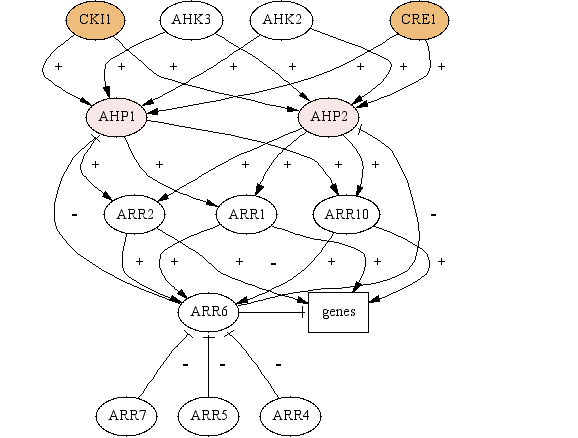
click to show details |
back to top
|
| Education
powerpoint presentation
Plant signaling
back to top
|
| Review
Müller, B.and Sheen, J. 2007. Advances in Cytokinin Signaling.
Science 318:68-69 PDF
Rolland, F., Moore, B., and Sheen, J. 2002. Plant sugar sensing
and signaling. Plant Cell 14: S185-S205. PDF
Sheen, J. 2002. Phosphorelay and transcription control in
cytokinin signal transduction. Science 296: 1650-1652.PDF
Hwang, I, Chen, H.-C. and Sheen. J. 2002. Two-Component Signal
Transduction Pathways in Arabidopsis. Plant Physiol. 129:
500-515. PDF
Sheen,
J. 2001. Signal transduction in maize and Arabidopsis
mesophyll protoplasts. Plant Physiol. 127:1466-1475. PDF
Tena,
G., Asai, T., Chiu, W.-L., and Sheen, J. 2001. Plant mitogen-activated
protein kinase signaling cascades. Curr. Opin. Plant Biol.
4:392-400 PDF
back to top |
|
| Articles
Müller, B. and Sheen, J. 2008.
Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094-1097 PDF SUPP
Müller, B. and Sheen, J. 2007. Arabidopsis Cytokinin Signaling Pathway.
Sci. STKE 2007 407 cm5 PDF
Müller, B. and Sheen, J. 2007. Cytokinin Signaling Pathway.
Sci. STKE 2007 407 cm4 PDF
Kim, H.J., Ryu, H., Hong, S.H., Woo, H.R., Lim, P.O., Lee, I.C.,
Sheen, J., Nam, H.G. and Hwang, I. 2006. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis.
PNAS 103(3): 814-819 PDF
Moore, B., Zhou, L.,Rolland, F., Hall, Q., Cheng, W.-H., Liu,
Y.-X., Hwang, I., Jones, T., Sheen, J. 2003. Role of the Arabidopsis
Glucose Sensor HXK1 in Nutrient, Light, and Hormonal Signaling.
Science 14 332-336 Abstract
Full Text
SUPP.
Hwang,
I. and Sheen, J. 2001. Two-component circuitry in Arabidopsis
cytokinin signal transduction. Nature 413(6854):383-9 PDF
Kovtun,
Y., Chiu, W.-L. Tena, G., and Sheen, J. 2000. Functional
analysis of oxidative stress-activated MAPK cascade in plants.
PNAS. 97: 2940-2945.
Jang,
J., Fujioka, S., Tasaka, M., Seto, H., Takatsuto, S.,
Ishii, A., Aida, Yoshida, S., Sheen, J. 2000. A critical role
of sterols in embryonic patterning and meristem programming
revealed by the fackel mutants of Arabidopsis thaliana. Genes
& Dev. 14: 1485-1497.
Kovtun,
Y., Chiu, W.-L. Zeng, W. and Sheen, J. 1998. Suppression
of auxin signal transduction by a MAPK cascade in higher plants.
Nature, 395: 716-720.
Chiu,
W.-L. , Niwa, Y, Zeng, W, Hirano, T., Kobayashi, H, Sheen,
J. 1996. Engineered GFP as a vital reporter in plants. Current
Biol. 6: 325-330.
-
back to top
|
|
|