 |
| MAPK Project
FUNCTIONAL ANALYSIS OF PLANT
MAPK CASCADES IN STRESS AND HORMONAL SIGNALING
  Supported
by the NSF DBI Program and the NIH Supported
by the NSF DBI Program and the NIH
Summary
| 
|
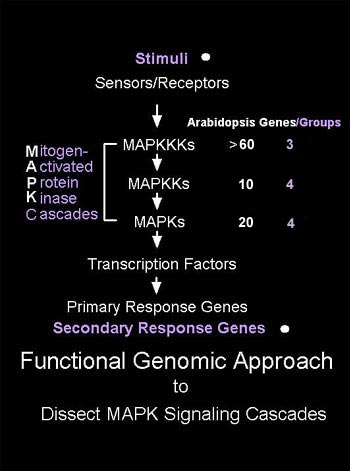
Mitogen-activated
protein kinase (MAPK) cascades are evolutionarily conserved
signaling modules with essential regulatory functions in
eukaryotes, including yeasts, worms, flies, frogs, mammals,
and plants. Numerous studies have shown that plant MAPKs
are activated by abiotic stresses, pathogens and pathogen-derived
elicitors, and plant hormones. The Arabidopsis genome and
EST sequencing projects have revealed large gene families
encoding MAPKs and their immediate upstream regulators,
MAPKKs and MAPKKKs. However, little is known about the constitution
of plant MAPK cascades and the specific roles that particular
MAPK cascade genes play in particular plant signal transduction
pathways. We are using a comprehensive approach based on
genomic information to generate a set of MAPK-, MAPKK-,
and MAPKKK-related clones in conjunction with transient
expression analysis to determine the function of all Arabidopsis
MAPK cascade genes involved in essential plant signaling
pathways. Since the functions of MAPK cascades in plant
signal transduction pathways are likely conserved, our studies
using the Arabidopsis genome resources will have broad implications
and applications in other plant species. |
|
| 
|
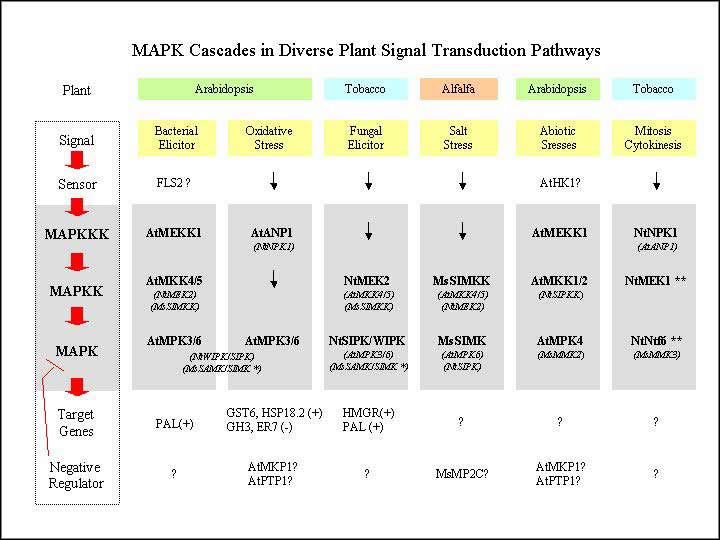
Our current studies
focus on five MAPK cascades, including those involved in
osmotic stress responses, Flg22 peptide-mediated defense
responses, the ethylene pathway, and auxin and H2O2
signaling. The overall strategy involves the identification
and cloning of the complete set of Arabidopsis genes encoding
MAPKs, MAPKKs, MAPKKKs, and relevant protein phosphatases.
Transient expression of epitope-tagged MAPKs, MAPKKs, and
MAPKKKs will be used to determine which MAPK cascades respond
to particular abiotic or biotic stress or hormone signals
in Arabidopsis and maize protoplasts. Constitutively active
and dominant negative forms of these MAPKKKs and MAPKKs
will be used to identify specific reporter genes associated
with particular signaling pathways. DNA array technology
will be used to analyze MAPK-related gene expression patterns,
and to reveal new downstream targets and crosstalk between
specific MAPK cascades. Identification of Arabidopsis knockout
mutants corresponding to key MAPK-related regulatory genes
will be initiated. Transgenic Arabidopsis, maize and soybean
plants that overexpress particular engineered MAPKKK and
MAPKK genes will be generated. The transgenic plants will
be tested for the exhibition of agronomically useful traits.
|
|
| |
The experimental
design is based on Arabidopsis and maize protoplast technology
developed in the Sheen and Ausubel laboratories. The transient
nature of the protoplast systems allows direct functional
analysis of plant genes at an unprecedented rate and at
relatively low cost. This approach takes full advantage
of the Arabidopsis genome infrastructure that is being created
as a part of the NSF Plant Genome Research Program, which
includes genome and EST sequencing, DNA microarray gene
expression technology, and knockout mutant libraries. The
experimental approaches that we are using are especially
powerful in unraveling the functions of genes that are difficult
to tackle by traditional genetic and biochemical approaches
due to redundancy, lethality or low levels of expression.
The elucidation and manipulation of MAPK cascades in plants
will reveal fundamentally important signaling processes
and provide new tools for crop improvement in stress tolerance,
disease resistance, and yield enhancement. |
|
back to top |
Signaling
Pathway Illustrations
Click to show details
back to top
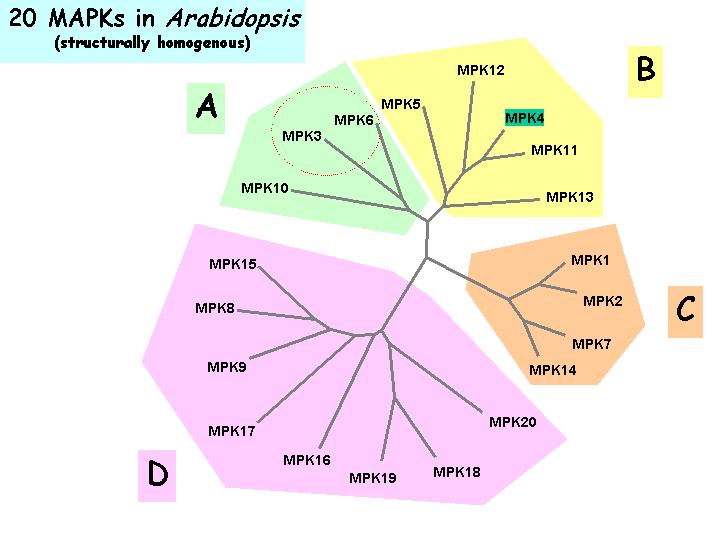
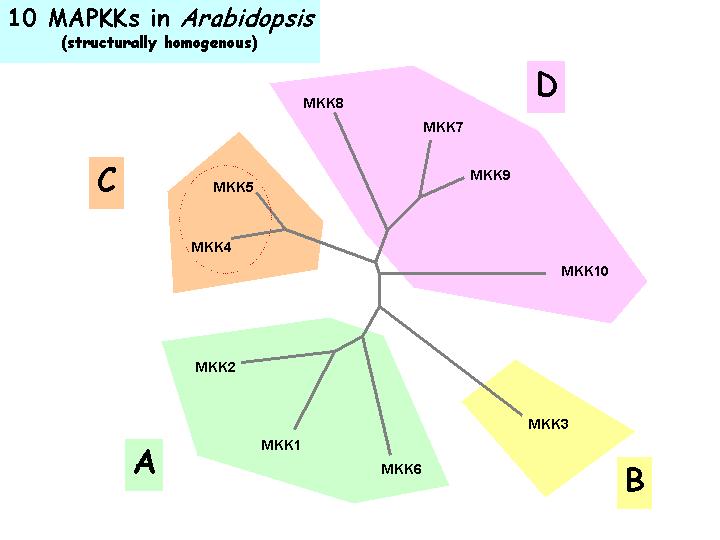
Dendrogram Tree Illustrations
Click to show details
|
 |
|
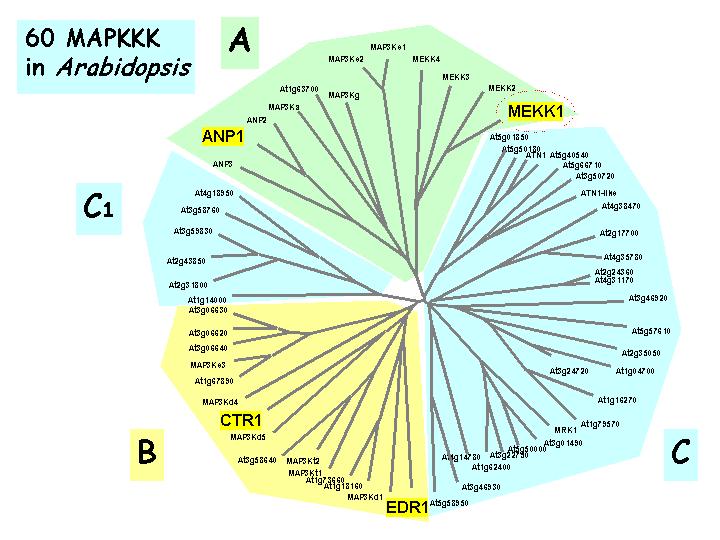
| Relatedness of Arabidopsis MAPKs |
Relatedness of Arabidopsis MAPKKs |
Relatedness of Arabidopsis MAPKKKs |
back to top
Publications |
| Reviews
Yoo, S-D., Cho, Y. and Sheen, J. 2009.
Emerging connections in the ethylene signaling network.
Trends in Plant Science 14 in press PDF
Boudsocq, M. and Sheen, J. 2009.
Calcium Sensing and Signaling. In: Abiotic Stress Adaptation in Plants: Physiological, Molecular and Genomic Foundation (A.Pareek, S.K. Sopory, H.J. Bohnert, Govindjee, eds). Springer, Dordrecht, The Netherlands: Springer PDF
Sheen, J., He, P., Shan, L., Xiong, Y., Tena, G., Yoo, S., Cho, Y., Boudsocq, M., and Lee, H. 2008.
Signaling specificity and complexity of MAPK cascades in plant innate immunity. Biology of Molecular Plant-Microbe Interactions. Chapter 86. PDF
Baena-González, E. and Sheen, J. 2008.
Convergent energy and stress signaling.
Trends in Plant Science 13:474-482 PDF
Müller, B. and Sheen, J. 2007.
Advances in Cytokinin Signaling.
Science 318:68-69 PDF
Shan, L., He, P., and Sheen, J. 2007. Intercepting Host MAPK Signaling Cascades by Bacterial Type III Effectors.
Cell Host & Microbe 1:167-174 PDF
He, P., Shan, L., and Sheen, J. 2007. Elicitation and Suppression of Microbe-Associated Molecular Pattern-Triggered Immunity in Plant-Microbe Interactions.
Cellular Microbiology 9:1385-1396 PDF
Yoo, S.-D., Cho, Y.-H. and Sheen, J. 2007. Arabidopsis Mesophyll Protoplasts: A Versitile Cell System for Transient Gene Expression Analysis.
Nature Protocols 2:1565-1572 PDF
Sheen, J. and He, P. 2007. Nuclear Actions in Innate Immune Signaling.
Cell 128:821-822 PDF
Shan, L., He, P., and Sheen, J. 2007. Endless Hide-and-Seek:
Dynamic Co-evolution in Plant-Bacterium Warfare. J.Integr. Plant Biol.
49(1):105-111 PDF
Hamel, L.-P., Nicole, M.-C., Sritubtim, S.,
Morency, M.-J., Ellis, M., Ehlting, J.,
Beaudoin, N., Barbazuk, B., Klessig, D., Lee, J.
Martin, G., Mundy, J., Ohashi, Y., Scheel, D., Sheen, J.,
Xing, T., Zhang, S., Seguin, A. and Ellis, B.E. 2006. Ancient signals:
comparative genomics of plant MAPK and MAPKK gene families : Trends in Plant Science
11(4): 192-198 PDF
Sheen, J. and Kay, S. 2004. Exploring new functions and actions of regulatory molecules.
Current Opinion in Plant Biology Volume 7: 487-490 PDF
Leon, P. and Sheen, J. 2003. Sugar and hormone connections.
Trends in Plant Science 8(3): 110-116 PDF
Ichimura, K., Tena, G., Henry, Y., Zhang, S., Hirt, H.,
Ellis, B. E., Morris, P. C., Wilson, C., Champion, A., Innes,
R. W., Sheen, J., Ecker, J. E., Scheel, D., Klessig, D. F.,
Machida, Y., Mundy, J., Ohashi, Y., Kreis, M., Heberle-Bors,
E., Walker, J. C. and Shinozaki, K. 2002. Mitogen-activated
protein kinase cascades in plants: a new nomenclature. Trends
in Plant Science 7: 301-308 PDF
Sheen, J. Signal transduction in maize and Arabidopsis mesophyll
protoplasts. Plant Physiol. 127:1466-1475 (2001).
PDF
Tena,
G., Asai, T., Chiu, W.-L., and Sheen, J. Plant mitogen-activated
protein kinase signaling cascades. Curr. Opin. Plant Biol.
4:392-400 (2001) PDF
back to top
|
| Articles
Shan, L., He, P., Li, J., Heese, A., Peck, S.C., Nürnberger, T., Martin, G.B. and Sheen, J. 2008.
Bacterial Effectors Target the Common Signaling Partner BAK1 to Disrupt Multiple MAMP Receptor Signaling Complexes and Impede Plant Immunity.
Cell Host & Microbe 4:17-27 PDF SUPP
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S.-Y., Cutler, S.R., Sheen, J., Rodriguez, P.L. and Zhu, J.-K. 2009.
In vitro reconstitution of an abscisic acid signaling pathway. Nature. 462: 660-664. PDF SUPP
Müller, B. and Sheen, J. 2008.
Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis.
Nature 453:1094-1097 PDF SUPP
Yoo, S-D., Cho Y-H., Tena G., Xiong. Y. and Sheen, J. 2008.
Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling.
Nature 451:789-795 PDF SUPP
Chen, Z., Agnew, J.L., Cohen, J.D., He, P., Shan, L., Sheen, J. and Kunkel, B.N. 2007.
Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology.
PNAS 104:20131-20136 PDF
Xiao, F., He, P., Abramovitch, R.B., Dawson, J.E., Nicholson, L.K., Sheen, J. and Martin, G.B 2007.
The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants.
The Plant Journal 52:595-614 PDF
Cho, Y.-H. and Yoo, S.-D. 2007.
ETHYLENE RESPONSE 1 Histidine Kinase Activity of Arabidopsis Promotes Plant Growth.
Plant Physiology 143: 612-616 PDF
Baena-Gonzalez, E., Rolland, F., Thevelein, J.M. and Sheen, J. 2007.
A central integrator of transcription networks in plant stress and energy signalling.
Nature 448: 938-943 PDF SUPP
He, P., Shan, L., Lin, N.-C., Martin, G.B.,
Kemmerling, B., Nurnberger, T., and Sheen, J.> 2006.
Specific Bacterial Suppressors of MAMP Signaling Upstream
of MAPKKK in Arabidopsis Innate
Immunity. Cell 125: 563-575 PDF SUPP
Shou, H., Bordallo, P., Fan, J.-B.
Yeakley, J.M., Bibikova, M., Sheen, J., and Wang, K. 2004.
Expression of an active tobacco mitogen-activated protein
kinase kinase kinase enhances freezing tolerance in transgenic
maize. PNAS 101(9): 3298-3303 PDF
Moore, B., Zhou, L.,Rolland, F., Hall, Q., Cheng, W.-H., Liu, Y.-X., Hwang, I., Jones, T., and
Sheen, J. 2003. Role of the Arabidopsis Glucose Sensor HXK1 in
Nutrient, Light, and Hormonal Signaling. Science 300:
332-336 Abstract
Full
Text SUPP.
Cheng, W.-H., Endo, A., Zhou, L., Penney, J.,
Chen, H.-C., Arroyo, A., Leon, P., Nambara, E.,
Asami, T., Seo, M., Koshiba, T., and Sheen, J.
2002. A Unique Short-Chain Dehydrogenase/Reductase in
Arabidopsis Glucose Signaling and Abscisic Acid Biosynthesis
and Functions. Plant Cell 14 2723-2743 PDF
Asai,
T., Tena, G., Plonikova, J., Willmann, M., Chiu, W.-L.,
Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J.
MAP kinase signaling cascade in Arabidopsis innate immunity.
Nature 415:977-983 (2002)
PDF
Asai, T., Stone, J. M., Head, J.E., Kovtun, Y., Yorgey, P.,
Sheen, J., and Ausubel, F.M. Fumonisin B1-induced cell death in
Arabidopsis protoplasts requires jasmonate-, ethylene-, and
salicylate-dependent signaling pathways. Plant Cell
12: 1823-1835 (2000).
Kovtun,
Y., Chiu, W.-L. Tena, G., and Sheen, J. Functional analysis
of oxidative stress-activated MAPK cascade in plants. PNAS
97:2940-2945 (2000)
Kovtun,
Y., Chiu, W.-L. Zeng, W. and Sheen, J. Suppression of
auxin signal transduction by a MAPK cascade in higher plants.
Nature 395: 716-720 (1998).
Chiu,
W.-L. , Niwa, Y, Zeng, W, Hirano, T., Kobayashi, H, and Sheen,
J. Engineered GFP as a vital reporter in plants. Current
Biol. 6: 325-330 (1996).
Sheen,
J., Hwang, S, Niwa, Y., Kobayashi, H., and Galbraith, D.W.
Green-fluorescent protein as a new vital marker in plant cells.
Plant J. 8: 777-784 (1995).
back to top |
|
|