| Line ID | Insert Site | Gene Length | Insert Site Feature | E-value | Forward Primer | Reverse Primer |
| SAIL_283_E11 | 1452 | 2624 | exon3 | 3.00E-16 | TGGAGCGCCGGTGTTATCTT | GTGGAGACTAAAGGGCTACAA |
| SAIL_187_E08 | 2739 | 2624 | exon7 | 0 | ACATCGCGGAGCTTAAAATCG | GTGTTGGGAGGAGCAGAACT |
|
Please note that: - SALK snapshots are updated on a monthly basis. - Information regarding the SAIL/GARLIC lines were based on A. Sessions et. al. (The Plant Cell, Vol. 14, 2985–2994, December 2002). - SAIL/GARLIC line insert site features were determined based on published insertion positions and gene feature tables obtained from TAIR. |
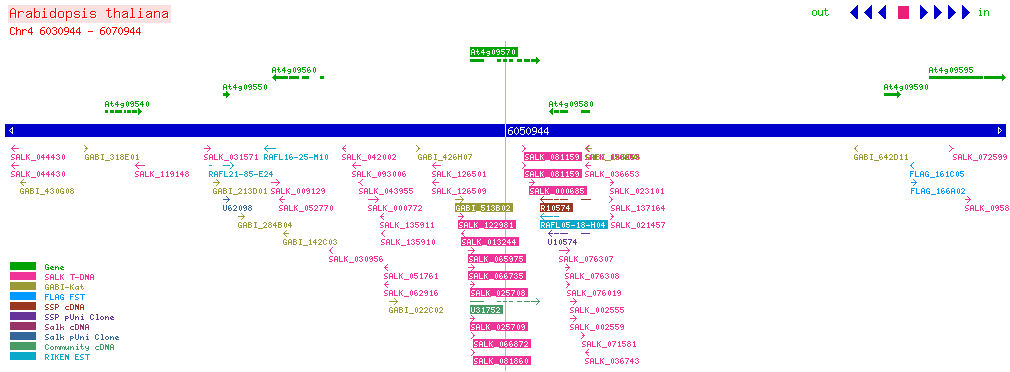
| CPK4 At4g09570 | |||||||||||||||||||||
| SALK: click here for updated view | |||||||||||||||||||||
 | |||||||||||||||||||||
| SAIL: | |||||||||||||||||||||
|
| CPK6(3) At2g17290 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SALK: click here for updated view | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
.gif) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SAIL: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| CPK26 At4g38230 | ||||||||||||||
| SALK: click here for updated view | ||||||||||||||
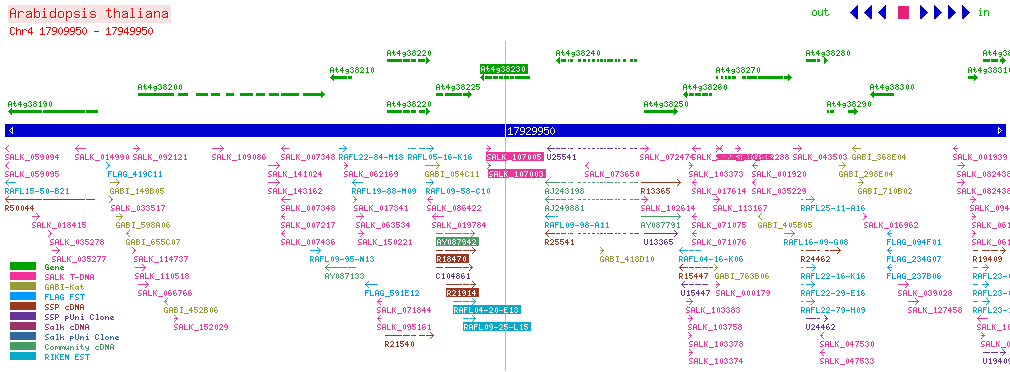 | ||||||||||||||
| SAIL: | ||||||||||||||
|
| CPK25 At2g35890 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SALK: click here for updated view | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
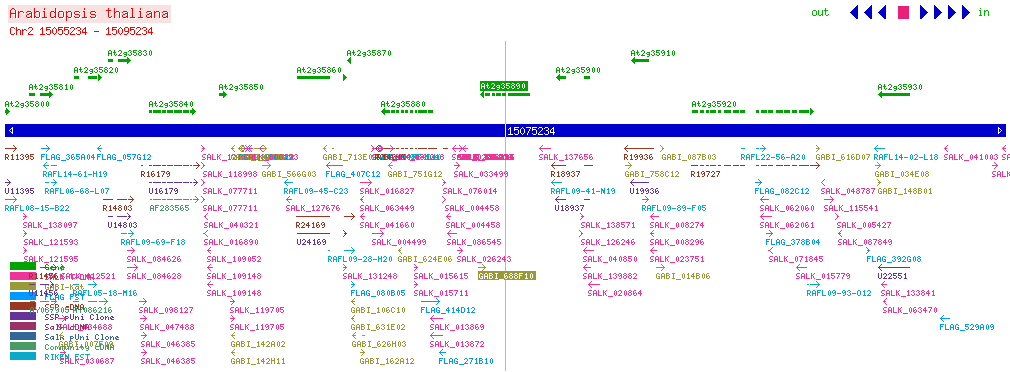 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SAIL: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| CPK11(2) At1g35670 | |||||||||||||||||||||
| SALK: click here for updated view | |||||||||||||||||||||
.gif) | |||||||||||||||||||||
| SAIL: | |||||||||||||||||||||
|
| CPK5 At4g35310 | ||||||||||||||||||||||||||||
| SALK: click here for updated view | ||||||||||||||||||||||||||||
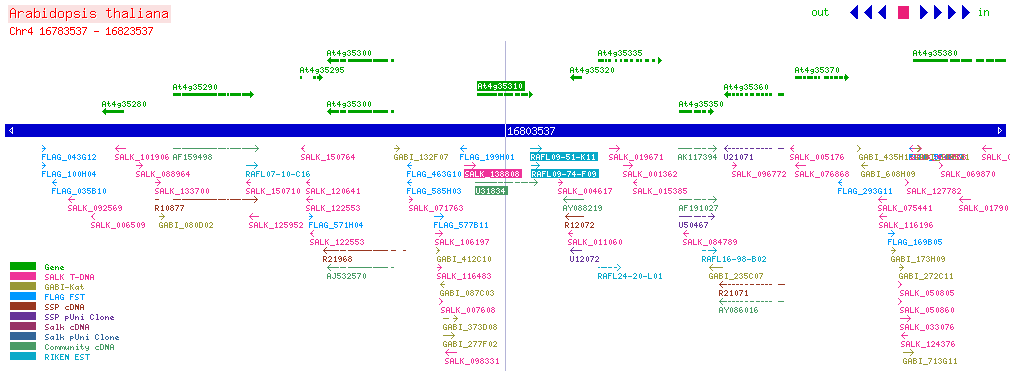 | ||||||||||||||||||||||||||||
| SAIL: | ||||||||||||||||||||||||||||
|
| CPK2 At3g10660 | |||||||||||||||||||||||||||||||||||
| SALK: click here for updated view | |||||||||||||||||||||||||||||||||||
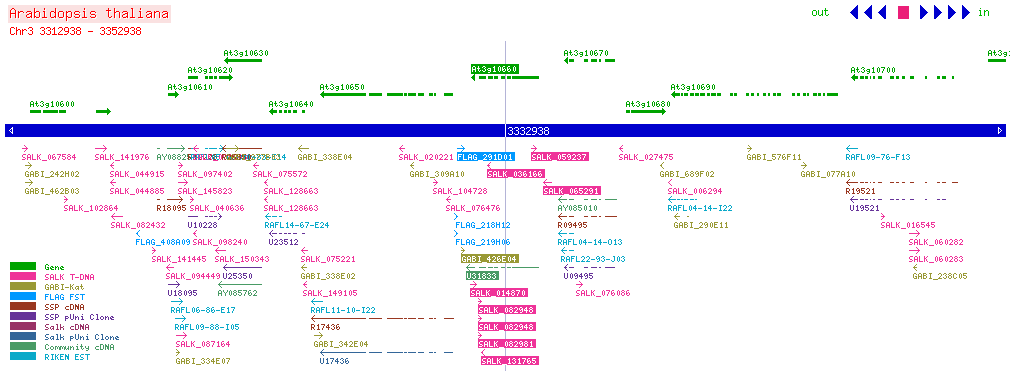 | |||||||||||||||||||||||||||||||||||
| SAIL: | |||||||||||||||||||||||||||||||||||
|
| CPK12(9) At5g23580 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SALK: click here for updated view | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
.gif) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SAIL: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| CPK1(AK1) At5g04870 | |||||||||||||||||||||||||||||||||||
| SALK: click here for updated view | |||||||||||||||||||||||||||||||||||
.gif) | |||||||||||||||||||||||||||||||||||
| SAIL: | |||||||||||||||||||||||||||||||||||
|
| CPK20 At2g38910 | |||||||||||||||||||||||||||||||||||||||||||||||||
| SALK: click here for updated view | |||||||||||||||||||||||||||||||||||||||||||||||||
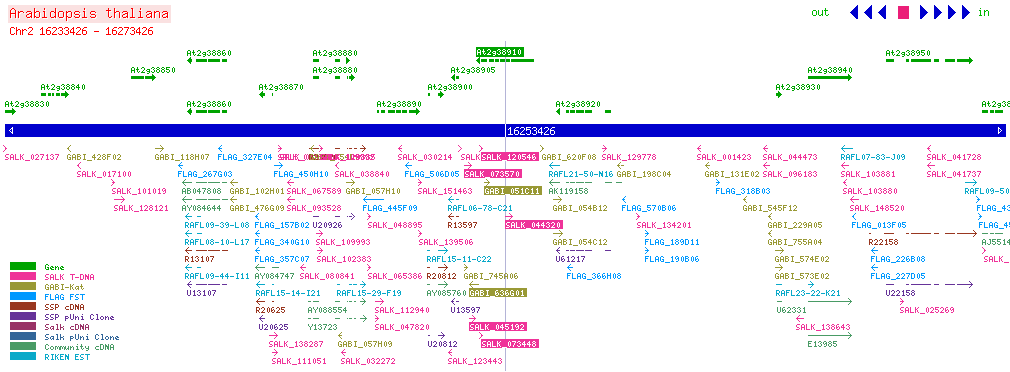 | |||||||||||||||||||||||||||||||||||||||||||||||||
| SAIL: | |||||||||||||||||||||||||||||||||||||||||||||||||
|