A Practical Evaluation of
Promoter Analysis and Search Tools:
How to Identify New & Important Cis-Elements
Alex Kazberouk & Mike Zhang
Summer Students 2005
Sheen Lab
MGH/HMS
Introduction
After the identification of co-regulated genes experimentally and computationally (e.g., microarrays, GeneChips), it is possible to use various computer programs to search their promoter regions for overrepresented motifs that may be cis-elements responsible for the regulation of the genes. It may also be useful to analyze the promoters to look for known cis-elements and to study the organization of and relationship between these putative cis-elements. This resource is designed to provide an overview and a brief evaluation of various bioinformatics tools useful for promoter analysis and cis-element searches for beginners like us. The emphasis is to explore useful tools for the analysis of Arabidopsis gene promoters.
The Fasta Format
Sequence Bulk Download and Analysis from TAIR
RSA-Tools – Retrieve Sequence
Motif Search Tools
Result Explanation
Weight matrices
Sequence logos
Promoter Searches
Motiffinder from TAIR
Weeder Web
MotifSampler
GeneSprings
MEME
TAIR Pattern Match
Genomatix
BioProspector
Improbizer
Toucan 2
Regulatory Sequence Analysis Tools
Databases Useful for Promoter Analysis
AGRIS
AthaMap
AtProbe
DoOP
PlantCare
PlantProm DB
Place
Transfac
Miscellaneous
Final Word
The Fasta Format
Most promoter analysis tools require that the sequence used for input is in Fasta format.
The format is:
>Description of sequence
Sequence itself
The description can be anything you want and is not looked at by programs.
The sequence should use the following codes:
A = adenosine M = A C (aMino)C = cytidine S = G C (Strong)G = guanine W = A T (Weak)T = thymidine B = G T CU = uridine D = G A TR = G A (puRine) H = A C TY = T C (pYrimidine) V = G C AK = G T (Keto) N = A G C T (aNy) - = gap of indeterminate length
For more information on Fasta see http://www.ncbi.nlm.nih.gov/BLAST/fasta.shtml
Two examples of Arabidopsis gene promoter sequences in Fasta:
>AT2G19190 5' sequence, length=500 This is an example of a promoter sequence in FastaTGATTCCTAAAAAAATATACAAACTATTGGGAGTTGTGAGATTTTTTATATCAGTGTTGGTCTCTTTACATTTGTGATGTGGTGTTATAGCATATATAGTAATAAACTCAAAAGGAAATTAGATGTGTTTTGACCATTTATTAAAATGAACCTTTTCTTGTCAAACATTTGAAAAATACTAGTTTTTTTTTTTGGCAACGTTGTAAATAATAGTTAAAAATAGATTTTAAGTCTCGTTTTTTTATGCATATAGTTTCATTCGCTTTATTAGACTCAAATATACTTTTAATTAAATTTTGCAGAGAATTAAAGGTAATCATTTGCCAAGGAAAAACCATGCAAATATGCAATAAGTAGAAATAATGTTAATGAGAGTAAGCGTTGACATATATTACGTCCTGGTCCGAACATTCTTAAAGTTGCGTAACACTAATAACCTTAGAAGATGGTTGGTTGACTATCAACATCTTATTGACCAAATGTTTTTTTTTTTTTAATTA >AT5G48430 5' sequence, length=500 Any sort of useful information can go hereGGTTATCATATCCTAGAAAACTTTCAAATCAGTGATTATCACATTCCATATTCAAACTATTGCTATTTATTTATTTCAATATATCTATATATATAATATTAATTGTCAACAAACATATAAAACCACAAGTAGTGATGCTAGTTGAAGTTTTTTCTTTTTCTTATTAACGTATAAGTCTTTTGAGTCTATGACCATCTACATCCATCAGTAGTTTTTAAATTTATCTATCAACTTGACATACATATAGGACTGGCTTTGCCTAAATGGTTGGCCCTCCTATGTCAACTTGATATACATATAGTAATCTTTTAAAATATATATATCTAGTTGTTCTGGTCGGACCGTATTATTGTGTGTACAAGTTCGTACATATTAACGTGTGTGATGTCTATATGTTAAAATGTCAACTCTAATTAGCTATATATACGTTCTTAATTCTCCTAGTAATTATATCCATATCTTAAAACTTTCCAACAATCTTAAACACAAACACGCAAAAG
It is much easier to use computer programs to convert your data to Fasta rather than to do it by hand. After getting the format, store it in Notepad or WordPad, not Word as Word may disrupt formatting.
See below for programs that provide sequences in Fasta:
Sequence Bulk Download and Analysis from TAIR (The Arabidopsis Information Resource)
http://www.arabidopsis.org/tools/bulk/sequences/index.jsp
1.) Paste in a list of genes, separated by commas, spaces, or carriage returns:
At2g19190
At5g48430
At2g24570
or
At2g19190, At5g48430, At2g24570
2.) In the dataset, select Upstream 500/1000/3000
3.) Make sure output format is set to Fasta and get the sequences
Output:
>AT2G19190 5' sequence, length=500 [CHR 2 START 8337027 END 8337526] REVERSETGATTCCTAAAAAAATATACAAACTATTGGGAGTTGTGAGATTTTTTATATCAGTGTTGGTCTCTTTACATTTGTGATGTGGTGTTATAGCATATATAGTAATAAACTCAAAAGGAAATTAGATGTGTTTTGACCATTTATTAAAATGAACCTTTTCTTGTCAAACATTTGAAAAATACTAGTTTTTTTTTTTGGCAACGTTGTAAATAATAGTTAAAAATAGATTTTAAGTCTCGTTTTTTTATGCATATAGTTTCATTCGCTTTATTAGACTCAAATATACTTTTAATTAAATTTTGCAGAGAATTAAAGGTAATCATTTGCCAAGGAAAAACCATGCAAATATGCAATAAGTAGAAATAATGTTAATGAGAGTAAGCGTTGACATATATTACGTCCTGGTCCGAACATTCTTAAAGTTGCGTAACACTAATAACCTTAGAAGATGGTTGGTTGACTATCAACATCTTATTGACCAAATGTTTTTTTTTTTTTAATTA>AT5G48430 5' sequence, length=500 [CHR 5 START 19646339 END 19646838] REVERSEGGTTATCATATCCTAGAAAACTTTCAAATCAGTGATTATCACATTCCATATTCAAACTATTGCTATTTATTTATTTCAATATATCTATATATATAATATTAATTGTCAACAAACATATAAAACCACAAGTAGTGATGCTAGTTGAAGTTTTTTCTTTTTCTTATTAACGTATAAGTCTTTTGAGTCTATGACCATCTACATCCATCAGTAGTTTTTAAATTTATCTATCAACTTGACATACATATAGGACTGGCTTTGCCTAAATGGTTGGCCCTCCTATGTCAACTTGATATACATATAGTAATCTTTTAAAATATATATATCTAGTTGTTCTGGTCGGACCGTATTATTGTGTGTACAAGTTCGTACATATTAACGTGTGTGATGTCTATATGTTAAAATGTCAACTCTAATTAGCTATATATACGTTCTTAATTCTCCTAGTAATTATATCCATATCTTAAAACTTTCCAACAATCTTAAACACAAACACGCAAAAG>AT2G24570 5' sequence, length=500 [CHR 2 START 10446378 END 10446877] REVERSETTCTAATATTCCAAAGAAACAAAAAAAAATCAGCCCAATTGTTCATACAAATAAAAACAATTCATTACCTTTAATTTTAAATTTATTGACTTGAACTTGAAGACATAAGATACCTAATAAAAGAAAAAATAGATATGAGACTTTAAAAAAGCTTTATGATTTTCTTTAGACACCATCCTTTAATGTTTTTTATTTGACTTTTTGTTTCTTTGAAATTCCTTTACCACCATTTTCCCCAAATTCAAGTTTACGCACAATGATTCCTTTATTTTAAAGACACGATTATAAATTCTTGCTTTGCACAAAAGAAGACCCTACATATCTCACAACTCAAGGAGACCAAACTTTGATTACTTTATTCCATAGAAATCTTCAACTCAATCTCAGCCGTTAGATCTAAAGCACCGATTTGACTAAACTCCATCTTAAACCTACTCAACCGGTCACTCGGTCACACCCATAACCCCATATATCACGCCAACGCCATTCTTTTTTCTTCCThis program works only for Arabidopsis. It selects a given number of base pairs upstream from each gene (does not include 5’ UTR) based on the sequenced genome. Thus, it is possible that the sequence you get overlaps with the open reading frame or the flanking sequences of other genes. The program also provides the length, the location, and the direction of the sequence submitted.
RSA-Tools – Retrieve Sequence
1.) Select retrieve sequence in the top left of web site
2.) If the loading time is slow, select a different server. We found that the Canadian one is usually reliable and fast.
3.) Select the organism and paste in the list of genes separated by carriage returns:
At2g19190
At5g48430
At2g24570
4.) Select how far upstream you want to search using negative numbers (the default is from -1500 to 0, 1500 base pairs upstream)
5.) Select whether you want the output displayed on your screen, saved on their server, or e-mailed to you
Output:
>At2g19190 At2g19190; upstream from -300 to 0; size: 301; location: NC_003071.3 8336975 8337275 R; upstream neighbour: At2g19200 (distance: 3324)
AGTTTCATTCGCTTTATTAGACTCAAATATACTTTTAATTAAATTTTGCAGAGAATTAAA
GGTAATCATTTGCCAAGGAAAAACCATGCAAATATGCAATAAGTAGAAATAATGTTAATG
AGAGTAAGCGTTGACATATATTACGTCCTGGTCCGAACATTCTTAAAGTTGCGTAACACT
AATAACCTTAGAAGATGGTTGGTTGACTATCAACATCTTATTGACCAAATGTTTTTTTTT
TTTTAATTATAAAACAGTTGCTCATTGCTCTAGCCCAGAGAAAGCAGCTCAATTAAGTAA
>MJE7.6 At5g48430; upstream from -300 to 0; size: 301; location: NC_003076.4 19646338 19646638 R; upstream neighbour: 15238972 (distance: 819)
TCCATCAGTAGTTTTTAAATTTATCTATCAACTTGACATACATATAGGACTGGCTTTGCC
TAAATGGTTGGCCCTCCTATGTCAACTTGATATACATATAGTAATCTTTTAAAATATATA
TATCTAGTTGTTCTGGTCGGACCGTATTATTGTGTGTACAAGTTCGTACATATTAACGTG
TGTGATGTCTATATGTTAAAATGTCAACTCTAATTAGCTATATATACGTTCTTAATTCTC
CTAGTAATTATATCCATATCTTAAAACTTTCCAACAATCTTAAACACAAACACGCAAAAG
>At2g24570 At2g24570; upstream from -300 to 0; size: 301; location: NC_003071.3 10446301 10446601 R; upstream neighbour: At2g24580 (distance: 5705)
ACACGATTATAAATTCTTGCTTTGCACAAAAGAAGACCCTACATATCTCACAACTCAAGG
AGACCAAACTTTGATTACTTTATTCCATAGAAATCTTCAACTCAATCTCAGCCGTTAGAT
CTAAAGCACCGATTTGACTAAACTCCATCTTAAACCTACTCAACCGGTCACTCGGTCACA
CCCATAACCCCATATATCACGCCAACGCCATTCTTTTTTCTTCCAGTTTCGCTCTCTCAT
TCATCAAAAAAAACTTGCACATCTTCTCAGATCTTCAAGTTTCTCCTCTGGTTTCTCATC
(note, all the information before the actual sequence is on one line)
The program works for 255 organisms and is not limited to Arabidopsis. It takes into account nearby genes and does not overlap into the previous open reading frame. However, it is slower and takes longer to use. The output is more informative and gives the location, distance, size, direction, and nearest gene. The output includes the 5’ UTR and starts at the predicted start codon. However, we found that not all Arabidopsis gene promoter sequences are available.
Motif Search Tools
Result Explanation
In addition to giving simple results with possible motifs, many programs give results as position weight matrices or as sequence logos.
Weight matrices:
Blk1 A C G T
1 56.39 0.09 4.41 39.10
2 99.61 0.09 0.09 0.21
3 34.78 26.02 0.09 39.10
4 0.21 0.09 99.49 0.21
5 0.21 0.09 99.49 0.21
6 69.36 0.09 0.09 30.46
7 99.61 0.09 0.09 0.21
8 0.21 99.49 0.09 0.21
This is the simplest example of a weight position matrix where the frequency of each nucleotide occurring in each position in the sequence is shown as a percentage. Thus, the motif is AA_GGAAC. Other possible weight matrices may show the information as log (frequency/expected frequency) and will give negative numbers for underrepresented nucleotides and positive ones for overrepresented ones.
Sequence logos:
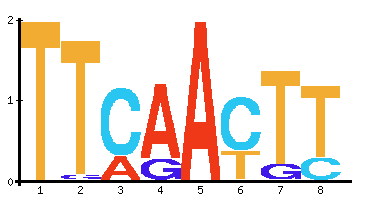
The four nucleotides can be represented as 2 sets of zeroes and ones (00, 01, 10, 11). The sequence logo displays this visually by showing the relative abundance of each nucleotide in the sequence based on the letter’s height. The maximum height is two.
Promoter Searches
Many programs exist to search for interesting and recurring motifs within putative regulatory sequences. Here are a few programs that might be useful:
Statistical Motif Analysis in Promoter or Upstream Gene Sequences:
Motiffinder from TAIR
http://www.Arabidopsis.org/tools/bulk/motiffinder
1.) Paste in a list of sequences to analyze in Fasta or paste in a list of genes whose promoters you want analyzed:
At2g19190
At5g48430
At2g24570
2.) If pasting a list of genes, select how far upstream you want to go
3.) Submit the data
Output:
|
ACCATC |
3 |
5235 |
3/3 |
4670/28088 |
4.60e-03 |
AT2G19190 AT5G48430 AT2G24570 |
|
GATGGT |
3 |
5236 |
3/3 |
4671/28088 |
4.60e-03 |
AT2G19190 AT5G48430 AT2G24570 |
|
CAACTC |
4 |
5679 |
3/3 |
5009/28088 |
5.67e-03 |
AT2G19190 AT5G48430 AT2G24570 |
The program is by far the simplest to use but applies only to Arabidopsis. It only searches 6 base pair motifs and uses a very simple search algorithm. The program does not allow for discrepancies and does not try to align sequences. It is very quick and works well when there are many (100+) sequences, but should not be used for very few sequences. It often gives false positives, but is a good starting point.
The output gives the motif, the total number of occurrences, the total number of sequences where it was found, how common the motif is in the genome, and the p-value for the motif. The results are listed by p-value with the lowest being on top. The results come in pairs, one for the forward and one for the reverse sequence.
Weeder Web:
http://159.149.109.16:8080/weederWeb/
1.) Enter your e-mail and paste the sequences in Fasta format
2.) Select your organism, the scan type, and the name of your job
3.) Click submit
Output:
The output is stored on their server and consists of a frequency matrix, a list of motifs, and a visual sequence logo:
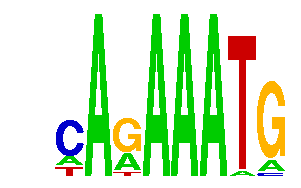
Weeder Web accepts 14 organisms, not just Arabidopsis. It uses a more sophisticated Weeder algorithm and usually gives good results. The program only accepts Fasta format and does not accept entries that are too long. It is better for fewer sequences. If the total input given is greater than 20k base pairs, only a quick run will be done. Quick runs take little time if the server is empty while a complete scan can take a day.
The output is easy to read. To ensure a likely positive, you should run the program several times as the results may be different. Usually, results which appear as the top few repeatedly are good candidates. For more information about interpreting output see: http://159.149.109.16:8080/weederWeb/output.html
MotifSampler:
http://www.esat.kuleuven.ac.be/~thijs/Work/MotifSampler.html
1.) Enter your e-mail address, name, and paste the sequences in Fasta format
2.) Select the organism and specify other parameters such as length and run number
3.) Run the search
Output:
The output is very similar to Weeder Web and also includes a frequency matrix and the occurrence of motifs. It also aligns the motifs with respect to their position on the sequences you input.
3 TTCAAyTT
|
|
|
Motif Sampler accepts many organisms including Arabidopsis. It uses a Gibbs sampling algorithm and usually gives good results. Tweaking the parameters can lead to many varied results. The results are different from run to run and the program should be run several times to ensure that a motif is likely to be a cis-element. The output is simple to read and is similar to Weeder Web. Other Motif Samplers that use the Gibbs sampling algorithm exist (ex: http://bayesweb.wadsworth.org/gibbs/gibbs.html). However, they tend to give similar results and are harder to use. If you need to tweak more parameters, the alternative programs may serve you better.
GeneSprings:
The program requires a subscription and should be installed on a laboratory computer.
1.) Load up GeneSprings and make sure that the Arabidopsis genome is the default one selected (ATH1)
2.) Add a list of genes either as data from an experiment or by hand. To add by hand, go to edit/edit gene list. There, paste a list of genes into the boxes on the right:
At2g19190
At5g48430
At2g24570
If the program does not recognize the gene, they will light up red. Add the genes to your list and save the list where you want it to be.
3.) Go to tools/find potential regulatory sequences, select your gene list, select number of discrepancies allowed, and run the search
Output:
AAGTC 36/52 0.023 38.19% 37.11% 19.86 5.68e-6 4096
AAAGTC 26/52 0.0043 15.64% 18.46% 9.602 2.63e-7 16384
TCAACTC 11/52 0.155 3.451% 3.69% 1.919 2.56e-6 65536
The sequence AAGTC was observed upstream of 36 out of the 52 genes in the gene list called WRKY29. Upstream means from 10 to 500 bases upstream of the gene. Only exact matches were counted. This was compared to the frequency (0.371) of that sequence upstream of other ORFs in the genome ATH1 w. sequence. If the distribution of bases were random you would expect to see that sequence upstream of 0.382 of the genes. The probability that this particular sequence is that common due to chance is 5.68e-6. However since 4,096 tests were done, the false positive probability is really 0.023.
The program can run for any organism genome that you give it. The algorithm used is simple and the promoter analysis tool is only a very small part of a much larger program. Sometimes, the program crashes or has fatal errors forcing you to restart. Before looking for sequences, make sure that the p-value cut off is high enough as the program runs multiple times and the p-value tends to be high for all motifs. The N’s in regulatory sequences allow you to look for interrupted motifs (AGCnnnnnnGCAAT) but we haven’t found them very useful.
After finding a motif, it is good to “View Genes for Selected Rows” to see the flanking of the motif and more information about it described in understandable language. After finding an over-represented sequence motif, it is a good idea to search for it again with slightly different criteria (higher upstream, more possible discrepancies, etc) using the much faster specific sequence search.
MEME:
http://meme.sdsc.edu/meme/website/meme.html
1.) Submit your e-mail and the description of your sequences
2.) Paste in the sequences in Fasta format
3.) Specify the minimum and maximum width of each motif and the number of motifs to find
Output
The output consists of many parts including alignment data, a frequency matrix, and motif locations:
MOTIF 3 width = 50
sites = 6 llr = 209
E-value = 2.3e+003
|
|
A |
|
: |
2 |
3 |
3 |
a |
3 |
2 |
: |
2 |
3 |
: |
3 |
: |
a |
7 |
5 |
: |
: |
5 |
: |
2 |
a |
8 |
2 |
3 |
2 |
3 |
7 |
3 |
2 |
2 |
5 |
7 |
: |
: |
: |
2 |
3 |
: |
5 |
2 |
a |
: |
: |
2 |
5 |
8 |
a |
3 |
2 |
|
|
|
pos.-specific |
C |
|
8 |
5 |
: |
: |
: |
3 |
2 |
7 |
5 |
2 |
3 |
2 |
: |
: |
: |
: |
: |
5 |
3 |
: |
7 |
: |
: |
5 |
: |
7 |
3 |
: |
: |
7 |
3 |
5 |
2 |
: |
7 |
a |
3 |
: |
: |
: |
2 |
: |
7 |
7 |
5 |
2 |
: |
: |
2 |
8 |
|
|
probability |
G |
|
2 |
: |
2 |
: |
: |
: |
: |
3 |
2 |
3 |
: |
5 |
7 |
: |
2 |
: |
: |
: |
: |
3 |
2 |
: |
2 |
: |
: |
2 |
: |
: |
2 |
2 |
2 |
: |
2 |
a |
2 |
: |
3 |
: |
: |
2 |
7 |
: |
: |
2 |
: |
2 |
2 |
: |
5 |
: |
|
|
matrix |
T |
|
: |
3 |
5 |
7 |
: |
3 |
7 |
: |
2 |
2 |
7 |
: |
3 |
: |
2 |
5 |
a |
5 |
2 |
7 |
: |
: |
: |
3 |
7 |
: |
3 |
3 |
5 |
: |
3 |
: |
: |
: |
2 |
: |
2 |
7 |
a |
3 |
: |
: |
3 |
2 |
3 |
2 |
: |
: |
: |
: |
|
. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2.7 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||
|
|
|
2.4 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
2.2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
1.9 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
Information |
1.6 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
content |
1.4 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
(50.2 bits) |
1.1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
0.8 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
0.5 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
0.3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
|
|
|
0.0 |
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
C |
C |
T |
T |
A |
A |
T |
C |
C |
A |
T |
G |
G |
A |
A |
A |
T |
C |
A |
T |
C |
A |
A |
C |
T |
C |
A |
A |
T |
C |
C |
A |
A |
G |
C |
C |
C |
T |
T |
A |
G |
A |
C |
C |
C |
A |
A |
A |
G |
C |
|||||||||
|
|
consensus |
|
|
|
T |
A |
A |
|
C |
|
G |
|
G |
C |
A |
T |
|
|
T |
|
T |
C |
G |
|
|
|
T |
A |
|
C |
T |
A |
|
T |
C |
|
|
|
|
G |
A |
|
T |
|
|
T |
|
T |
|
|
|
A |
|
||||||||
|
|
sequence |
|
|
|
|
|
|
|
T |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
T |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||||||
MEME is commonly used to find motifs for many organisms although we have not found it very useful for our project yet. It is not specific to Arabidopsis and can be used for any organism. It is more suited to finding longer motifs and not short cis-elements, so you should specify motif length to be short as one of the parameters. It tends to pick up a lot of background noise and is somewhat difficult to use effectively. Along with MEME results, an extensive analysis of alignment data is sent to the user from MAST, a sister program. We found the MAST results harder to read and not very helpful in finding cis-elements. However, MEME overall can be used well with other programs especially for verification. There are published papers that used MEME to find long cis-elements.
TAIR Pattern Match:
http://www.Arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl
1.) Select nucleotide sequence and enter your putative motif
Ex: TCAACT
2.) Choose the sequence database upstream 500/1000/3000
3.) Start pattern search
Output:
|
Hit# |
Sequence name |
# of hits |
Hit |
Matching Positions |
Hit |
|
|
start |
end |
|||||
|
1 - 6 |
6 |
TCAACT |
28 |
33 |
||
|
TCAACT |
50 |
55 |
||||
|
TCAACT |
80 |
85 |
||||
|
TCAACT |
371 |
376 |
||||
|
TCAACT |
390 |
395 |
||||
|
TCAACT |
462 |
467 |
||||
The program searches for repeating short sequences through the entire Arabidopsis genome and finds places where they are most common. Thus, it is possible to search for a putative cis-element among the promoters of many genes and find other genes which can be regulated by it. This can be further validated by performing experiments to see if the motifs are important for gene regulation. However, this cannot be used to find new motifs, merely to check known ones. Thus, the program is of limited use.
Genomatix:
1.) Go to access and buy and pick “free of charge evaluation account”
2.) Proceed to follow directions to register making sure to select academic (rather than commercial) as your license.
The program developed in Germany is a massive database and set of analysis tools. However, it is a commercial product and costs thousands of euros. For academic users it offers a smaller limited free service that can be very useful and regenerates every month. There are many components to the program and they are described in details on:
http://www.genomatix.de/online_help/help_gems/faq.html
A few available tools are:
ElDorado: Provides very detailed information and maps of known functional genomic elements within a gene: promoters, enchancers, introns/exons, etc. For usage go to:
http://www.genomatix.de/online_help/help_eldorado/introduction.html
MatInspector: Provides very good information about all elements possibly involved in regulation of a gene. The information given is good for verification and also allows easy location of each element.
Bibliosphere: Allows data search and extraction of gene relation on gene databases and genome wide sequence analysis
Gene2Promoter and FrameWorker (under GEMS Launcher): Both can be used for the analysis of co-regulated genes and looking for promoters that might be involved.
For more, see:
http://www.genomatix.de/online_help/help_eldorado/Gene2Promoter_Intro.html
http://www.genomatix.de/online_help/help_gems/FrameWorker.html
Note: When using, don’t forget to specify organism/organism type. Results vary greatly. Also, keep track of the number of searches you are allowed per month on the free license.
Overall, the free parts of the program make it potentially very useful. While the paid sections would make it better, many other free tools are available online that can get the job done just as well. Thus, it is not necessary to spend thousands of euros for the program if you do not the funds.
BioProspector:
http://ai.stanford.edu/~xsliu/BioProspector/
1.) Click on MotifFinding on the left of the screen
2.) Specify your e-mail and paste in input sequences in Fasta Format
3.) Paste in a background model if you have one and submit the search
Output:
The highest scoring 5 motifs are:
Motif #1: (AGTTGACT/AGTCAACT)
******************************
Width (8, 0); Gap [0, 0]; MotifScore 3.084; Sites 16
Blk1 A C G T Con rCon Deg rDeg
1 92.75 0.26 6.40 0.60 A T A T
2 0.60 0.26 98.55 0.60 G C G C
3 0.60 0.26 0.26 98.89 T A T A
4 0.60 0.26 18.69 80.46 T A T A
5 0.60 0.26 98.55 0.60 G C G C
6 98.89 0.26 0.26 0.60 A T A T
7 43.60 55.55 0.26 0.60 C G M K
8 0.60 0.26 30.97 68.17 T A K M
The program is not specific to Arabidopsis and is thus somewhat limited in its usability for us. However, it can be used for further confirmation of the data. The output gives a list of possible motifs, their length, a matrix of sites and base pairs in the motif, and where the motif is found on each input sequence.
Because Arabidopsis is not one of the possible background models it is possible to make the results better by including several known Arabidopsis promoter sequences as a background model. This is used by the program to determine factors such as the expected base pair ratios. The default choice is to use the input data as a background and that usually works well by itself.
Improbizer:
http://www.mitoz.bcs.uwa.edu.au/improbizer/index.php
1.) Paste in sequence in Fasta format
2.) Modify any specifications necessary and paste in background sequences if you want better results
Output:
Converging..................................
7.1224 @ 589.79 sd 137.59 TTTCACGG
a 0.277 0.277 0.031 0.003 0.826 0.031 0.003 0.140
c 0.223 0.058 0.086 0.853 0.003 0.497 0.058 0.031
g 0.031 0.223 0.003 0.140 0.168 0.222 0.935 0.826
t 0.469 0.442 0.881 0.003 0.003 0.250 0.003 0.003
The program is not specific to Arabidopsis and uses a fairly old algorithm. All computation is done directly on the server and in real time, so no e-mail is necessary. The output provides the sequence and the matrix and later provides a list of all the sequences and a color-coded display of where each motif is located in each sequence.
This program uses a different algorithm and could potentially be useful for promoter analysis. However, there are many better and nicer programs that exist.
Toucan 2:
http://www.esat.kuleuven.ac.be/~saerts/software/toucan.php
1.) Download necessary Java applications to launch Toucan 2
2.) Follow directions outlined in tutorials on: http://www.esat.kuleuven.ac.be/~saerts/software/tutorial1/TOUCAN_Tutorial_Overview.html
Toucan 2 is a BioJava application designed for analysis of regulatory elements of higher organisms. However, it takes data from databases that do not have plant models. Thus, to enter sequences you need to create files with sequences in Fasta format and give them .fasta extensions (not .txt or .doc). These files can then be used to input sequences into the program to analyze. Also, when selecting backgrounds, it is better to use 3rd order plants as these tend to give the best results.
Toucan uses a variety of algorithms and gives a great deal of output that is described well on the tutorial website. The output includes a clear visual map of the input sequences with color coded putative cis-elements on them. The program is a very powerful tool, but is also difficult to use and not suited specifically for Arabidopsis. However, its many different approaches to looking for regulatory sequences (alignment, string searches, matrices, etc) make it useful.
Regulatory Sequence Analysis Tools:
1.) After getting sequences in Fasta format, use any of the pattern discovery applications on the left
2.) Follow instructions as necessary and choose e-mail output because some computations will take a long time
3.) Make sure to change organisms as necessary and use the default characteristics given by the website for a first run-through
Output:
|
seq |
identifier |
exp_freq |
occ |
exp_occ |
occ_P |
occ_E |
occ_sig |
ovl_occ |
rank |
test |
|
agtcaa |
agtcaa|ttgact |
0.0007669635693 |
14 |
0 |
2e-06 |
4.1e-03 |
2.38 |
0 |
1 |
right |
|
gtcaac |
gtcaac|gttgac |
0.0004262449032 |
8 |
0 |
0.00026 |
5.5e-01 |
0.26 |
0 |
2 |
right |
|
tgacca |
tgacca|tggtca |
0.0004532945152 |
8 |
0 |
0.00040 |
8.2e-01 |
0.09 |
0 |
3 |
right |
Note: The output looks fairly similar between different programs as only the algorithms used to look for cis-elements are different.
The website uses many different algorithms to search for possible motifs. It was built for simple genomes (prokaryotes and yeast) and does not work perfectly for Arabidopsis, despite the fact that Arabidopsis is one of the organism choices offered. The results are very abundant, but often contradict each other between the different programs found on the website. There are also many false positives. Thus, we recommend using the program at the end to verify results rather than starting by using it.
For simple searches, it is best to use oligo-analysis (words) as that gives good results quickly. However, to verify, run multiple programs and see if the results match.
Make sure to check the detailed tutorial section found in the miscellaneous category for useful tips on adjusting input and ideas on how to better interpret the output.
Useful Databases for Promoter Analysis
AGRIS:
http://Arabidopsis.med.ohio-state.edu/
AGRIS currently contains two databases, AtcisDB (Arabidopsis thaliana cis-regulatory database) and AtTFDB (Arabidopsis thaliana transcription factor database)
ATCISDB is a database of cis-elements on the promoters of all genes in the Arabidopsis genome. It is not complete and not experimentally verified, but it provides a clear visual of the promoter, the promoter sequence, and a list of all putative cis-elements. Its putative promoter sequences are somewhat different from those from TAIR because this d has incorporated the information of full-length cDNA sequences from RIKEN and SALK. It also has links to other databases and information about cis-elements. Finally, it provides the distance between putative cis-elements and the beginning of translation. It also contains a list of all currently known plant transcription factor binding sites at http://Arabidopsis.med.ohio-state.edu/AtcisDB/bindingSiteContent.jsp
ATTFDB is a database of transcription factors that can be searched or browsed. It is an easy way to find genes coding for various transcription factors. It also provides links to other databases such as TIGR for information about the TFs.
For more information see: http://www.biomedcentral.com/1471-2105/4/25
Davuluri R.V., Sun H., Palaniswamy S.K.,
Matthews N., Molina C., Kurtz M., Grotewold E.
AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource
of Arabidopsis cis-regulatory elements and transcription factors
BMC Bioinformatics. 2003 Jun 23;4(1):25.
AthaMap:
A genome-wide map of putative transcription factor binding sites in Arabidopsis thaliana
The website uses the sequenced genome and other databases such as Transfac to list possible putative transcription factor binding sites on all putative promoters in the Arabidopsis genome. It provides a visual of the promoter and gives links to published information about each transcription factor. However, AGRIS has more information, is easier to use, and provides a clearer visual. This may be useful for copy/pasting sequences of the promoter with cis-elements marked (as AGRIS does not allow that).
For more information see: http://www.pubmedcentral.gov/articlerender.fcgi?artid=308752
Steffens N.O. Galuschka C. Schindler M. Bülow L. Hehl R.
AthaMap: an online resource for in silico transcription factor binding
sites in the Arabidopsis thaliana genome. Nucleic Acids Res. 2004;32:D368–D372
AtProbe:
http://exon.cshl.org/cgi-bin/atprobe/atprobe.pl
The Arabidopsis thaliana promoter binding element database.
An aid to find binding elements and check data against the primary literature
The database is a very clunky list of tables with no real search algorithm. It lists experimentally determined transcription factors and experimentally determined instances of them occurring in Arabidopsis. Thus, it should be used for checking the data you have. It is similar to PlantProm DB, but is specific to Arabidopsis. It is simple to learn, to use, but does not have as much information.
DoOP: Databases of Orthologous Promoters:
A database containing orthologous clusters of promoters from Homo sapiens, Arabidopsis thaliana and other organisms
The database is very useful for matching sequences between different species and has many genes with sequenced promoters. However, it does not take full advantage of the sequenced genome. It has basic information about genes and allows matching promoters of similar genes between different organisms. There are many ways to look for promoters and the promoters are not experimentally determined. Although there are better programs to look for promoters, the ability of this database to match promoters between species may be useful.
For more information see: (http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=540051)
Barta E., Sebestyén,E., Pálfy,T.B., Tóth,G., Ortutay,C.P. and Patthy,L.
DoOP: Databases of Orthologous Promoters, collections of
clusters of orthologous upstream sequences from chordates and plants. Nucleic Acids Res., 33,
D86–D90.
PlantCare:
http://oberon.fvms.ugent.be:8080/PlantCARE/
Database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences
The database has many useful links to various programs involved in promoter analysis. It also allows searching sequences for putative cis-elements, but it does not offer access to the entire genome like AGRIS. Few genes are available. Very little information about the W-box could be found (only its sequence and a short description of its function). The search for CARE tool is useful for inputting sequences and then finding possible cis-elements on them.
For more information see: (http://nar.oxfordjournals.org/cgi/content/full/30/1/325)
Lescot M., Dehais,P., Thijs,G., Marchal,K., Moreau,Y., Van
de Peer,Y., Rouze,P. and Rombauts,S. (2002) PlantCARE, a database of plant cis-acting
regulatory elements and a portal to tools for in silico analysis of
promoter sequences. Nucleic Acids Res., 30, 325–327.
PlantProm DB:
http://www.softberry.com/berry.phtml?topic=plantprom&group=data&subgroup=plantprom
Database with annotated, non-redundant collection of proximal promoter sequences for RNA polymerase II with experimentally determined transcription start site(s) (TSS) from various plant species
The site is fairly new and does not have a very large database. Its only purpose is to give experimentally determined promoter sequences very close to the transcription start site. It covers many plant species but currently only has 63 promoters for Arabidopsis and does not cover all genes. If the database were larger, it would be useful for comparative analysis of genes. If the genes you are studying are in the database, it would be good to use the database as the data here is experimentally supported.
For more information see: (http://nar.oxfordjournals.org/cgi/content/full/31/1/114)
Shahmuradov I.A. Gammerman A.J. Hancock J.M. Bramley P.M. Solovyev V.V. PlantProm: a database of plant promoter sequences. Nucleic Acids Res. 2003;31:114–117.
Place:
http://www.dna.affrc.go.jp/PLACE/
Database of motifs found in plant cis-acting regulatory DNA elements, all from previously published reports. It covers vascular plants only.
Place contains a useful signal scan search where it is possible to enter short sequences (<5000) and it supplies all possible cis-elements in that sequence. There is also a small amount of information on all the cis-elements known so far. Place also provides information on articles about cis-elements.
However, it is probably better to use something such as AGRIS because AGRIS offers more information, visuals, and allows searching by gene rather than by entering sequences. The Place database is also fairly old and does not utilize the sequenced genomes of plants.
For more information see: http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=9847208
Higo, K., Y. Ugawa, M. Iwamoto, and T. Korenaga. 1999. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27:297-300.
Transfac:
http://www.gene-regulation.com/pub/databases.html#transfac
Database on eukaryotic transcription factors, their genomic binding sites and DNA-binding profiles. A commercial site.
Transfac is a very large database covering many eukaryotic genomes. Originally it was free to use, but recently it has become commercial and requires yearly payments. The free version requires a registration and has a bad interface. After using search, it is best to go to factors to get a list of transcription factors or to sites to search for bindings sites.
It is probably better and cheaper to use other databases than Transfac, although the database is up to date and woks well.
For more information on searching see:
http://www.gene-regulation.com/pub/databases/transfac/doc/relations.html
For more information on TRANSFAC see:
http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=12520026
Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, et al.
TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids
Res. 2003;31:374–378.
Miscellaneous
RSAT
RSAT has a collection of minor tools that may be useful when looking for promoters. In the other tools section of the site, there is a function that allows the conversion between different formats, such as Fasta, IG, raw, and Wconsensus (convert sequence). For making background models, there is a random sequence function. By selecting an organism and needed characteristics of a sequence, it is possible to generate random sequences to act as background. This can give better results on programs such as Improbizer. There are also tools of looking for a specific pattern/sequence within a larger sequence or within an entire genome of an organism. We did not find it very useful, but genome scale DNA pattern searches can be used like TAIR pattern match.
Final Word
Although there are many programs available, much still needs to be done by hand. The programs are not perfect and sometimes they miss similar motifs or miss the reverse complements of the motifs. Careful examination of possible motifs given by a program can offer new insights. No program is 100% certain to be right and multiple runs of multiple programs are necessary.
After extensive motif searches using different programs, we found it helpful to copy promoters in Fasta format into Word to generate a final presentation. We could use the Word search tool to find and highlight/change the font/color/size of cis-elements such as W-boxes or newly found possible cis-elements. The results should match at least partially with the data from promoter search programs and databases.
A very simple algorithm that compares two lists and finds common words (Kevin Chu/Sheen Lab has it) saved us a lot of manual work. Multiple lists could be compared much faster to find which genes matched among which lists.
Many of these programs are new and are constantly being updated. Hopefully, many flaws will soon be fixed. Looking for new programs that might be out there might help your search!
For any questions, e-mail
kazberouk@molbio.mgh.harvard.edu