Download protocols for...
|
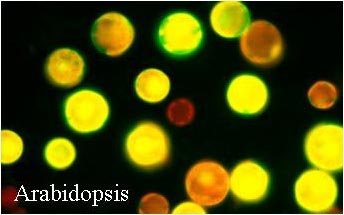
| A Transient Expression Assay Using Arabidopsis Mesophyll Protoplasts |
|
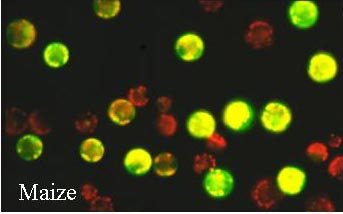
| A Transient Expression Assay Using Maize Mesophyll Protoplasts |
 Study
Signal Transduction in Arabidopsis and Maize Mesophyll Protoplasts
Study
Signal Transduction in Arabidopsis and Maize Mesophyll Protoplasts
| Plant protoplasts show physiological perceptions and responses to hormones, metabolites, environmental cues, and pathogen-derived elicitors, similar to cell-autonomous responses in intact tissues and plants. The development of defined protoplast transient expression systems for high throughput screening and systematic characterization of gene functions has greatly contributed to elucidating plant signal transduction pathways, in combination with genetic, genomic, and transgenic approaches. |
|||||||||||||||||||||||||||||
| INTRODUCTION The availability of mutants, transgenic plants, global gene expression profiles, and genomic sequences has offered invaluable opportunities for understanding organismal plant biology at the cellular and molecular level. Notably, molecular and genetic studies have discovered central components from receptors to transcription factors in diverse plant signal transduction pathways. Still, many missing links exist in the plant transduction pathways. |
|||||||||||||||||||||||||||||
| Tissue culture cell lines and transient expression assays have proven indispensable for rapid progress in signal transduction research in mammals. Analogous systems in plants have been established that use protoplast transient expression of parsley, maize, carrot, alfalfa, Arabidopsis, and tobacco suspension culture cells. These plant cell lines offer new opportunities to dissect signal transduction pathways involved in UV, ABA (abscisic acid), metabolite, ribosomal RNA, light, auxin, defense, and cell cycle regulation. |
|||||||||||||||||||||||||||||
| Compared with cell culture lines, the use of fresh tissues as protoplast sources offers unique advantages. For example, protoplasts isolated from plant tissues retain their cell identity and differentiated state. They also show high transformation efficiency with low maintenance. These freshly isolated protoplasts have proven to be physiological and versatile cell systems for studying a broad spectrum of plant signaling mechanisms underlying phytochrome, clock, auxin, GA, light, sugar, stress, auxin, H2O2, membrane transport, ABA, cytokinin, and cell death controls. Advances in novel protoplast assays will help make functional genomic and proteomic analyses of individual plant genes and their products a reality. They also provide a convenient and powerful tool for the cellular, genetic and molecular analysis and complementation of existing mutants. |
|||||||||||||||||||||||||||||
| We are providing two protoplast protocols detailing the preparation and selection of plant materials, protoplast isolation and incubation procedures, DNA transfection methods, and reporter gene assays, as well as key references and trouble-shooting tips. A more general introduction to the maize and Arabidopsis mesophyll protoplast systems is provided in a recent review (Sheen, Plant Physiol. 127: 1466-1475, 2001 PDF). A "FAQ" website focusing on the use of mesophyll protoplast systems has been posted. |
|||||||||||||||||||||||||||||
| The protocol has been streamlined and can be applied to different types of plant materials. The growth condition of plants seems to be the most critical factor for experimental reproducibility. Each lab may need to work out the best plant growth conditions for its setting. It usually takes some time to develop specific assays since these protoplasts are dynamic with numerous endogenous activities (a physiological system!). You may need to modify plant growth conditions or define specific physiological and developmental stages for protoplast isolation. It certainly helps if you think from a plant's point of view!! The protocol is simple and the approach is powerful. However, it only awards success to the scientists who are willing to take the time to master the system with patience and faith. Good luck with your experiments! |
|||||||||||||||||||||||||||||
| References for Arabidopsis : |
|||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||
| References for Maize: |
|||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||
| References for Rice: |
|||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||
| Registered already? |
| Registration form Fields marked with asterisk (*) are required
|